RNA Dynamics – A Bug or a Feature?
Donovan Chin, Director of Computational Drug Discovery, Arrakis

Though not nearly as famous as its double-stranded cousin, RNA is a fascinating molecule and its potential as a drug discovery substrate continues to unfold beyond merely transferring information from DNA into proteins. To perform its role in the cell, RNA must move, contort and adapt to a wide range of functional and regulatory requirements. RNA is dynamic. And understanding its dynamics can unveil new entry points for drug discovery and development.
Sequencing the human genome not only brought us new drug targets, but also a greater appreciation of RNA’s role in regulating the genome’s internal “wiring.” With this new understanding, RNA is an opportunity not only to tackle undruggable protein targets through the mRNAs that encode them (as we have discussed in past blogs), but also to open up a whole new world of drug targets by delving into complex RNA regulatory networks. To achieve this, we’ll need to grapple with RNA dynamics and wrestle them to the mat.
RNA Motions Revealed
The first x-ray crystallography structures of tRNA were disclosed in the 1970s and ribosome structure was elucidated by Steitz and others about 30 years later. While these structures are “snapshots” of RNA structure frozen in time, they laid the foundation for a detailed understanding of RNA dynamics. For example, ribozymes, small structures with complex folds and protein-like catalytic behavior, function as molecular machines. All these data hinted that RNA underwent functionally significant motion through conformational states that are a diverse collection of global domain-like motions and local base pair rearrangements.
Evidence of localized motion in protein function was emerging in parallel and contributed to the study of RNA dynamics. Since then, structural biology has shown very detailed motions of RNAs using NMR and, to an increasing extent, computation. In addition, there is greater appreciation of the role of various cell effectors (e.g., proteins, metabolites and other RNAs) to induce one functional RNA conformation over another.
Examples of the diverse interplay between RNA dynamics and function include microRNA processing, mRNA translation efficiency regulated by structures in the untranslated regions of that mRNA, riboswitch rearrangement upon ligand binding, RNA/protein interactions, and RNA splicing. The dynamics of these phenomena range from localized motions to large loop movements or helix unwinding, and their timescales are equally divergent. In many cases, functional responses can be related to either large-scale RNA domain motions or more nuanced localized dynamics of RNA.
Nucleotide base pairing and unpairing in the crowded environment of a cell is at the heart of RNA dynamics and ultimate functional outcome in cells. The pairing and unpairing is not haphazard, but rather a connected path that RNA must follow as it dances across its accessible conformational landscape. The challenge is to identify small molecule pockets within the changing conformational landscape of RNA that are functionally relevant. Regions with the right mix of non-canonical pairings and the more familiar Watson-Crick canonical base pairing may provide the first clue.
The Arrakis Edge
At Arrakis, our ambition is to create new therapeutics by modulating RNA dynamics using small molecules. To achieve this, we need to target pockets in RNA where small molecules can bind. It’s widely recognized that small molecules can bind along major or minor grooves, via base-pair complementarity, or by intercalation. But the real opportunity may lie in pockets fashioned by dynamic regions created by the non-canonical base pairings. Non-canonical base pairings are prevalent in RNA but are not as energetically favored as their canonical relatives (GC, AU), a fact that contributes to the dynamics. Small molecule pockets can occur, broadly speaking, near non-canonical base interactions or bulges in regions of mismatched base pairing. The confluence of these events can create context-dependent distortions in the three-dimensional fold of the RNA, resulting in a buried pocket surrounded by multiple non-covalent anchor points that are favorable for small molecule binding.
The figure below demonstrates our approach for scanning the RNA dynamic landscape and identifying likely ligand binding sites and binding modes. The approach yields a model that is consistent with a variety of experimental data, including our PEARL-seqTM platform and chemistry structure-activity relationships. As RNA is dynamic, so are the pockets. When they arise, these pockets may provide multiple opportunities to drug specific RNAs. The analogy in proteins is allosteric binding.
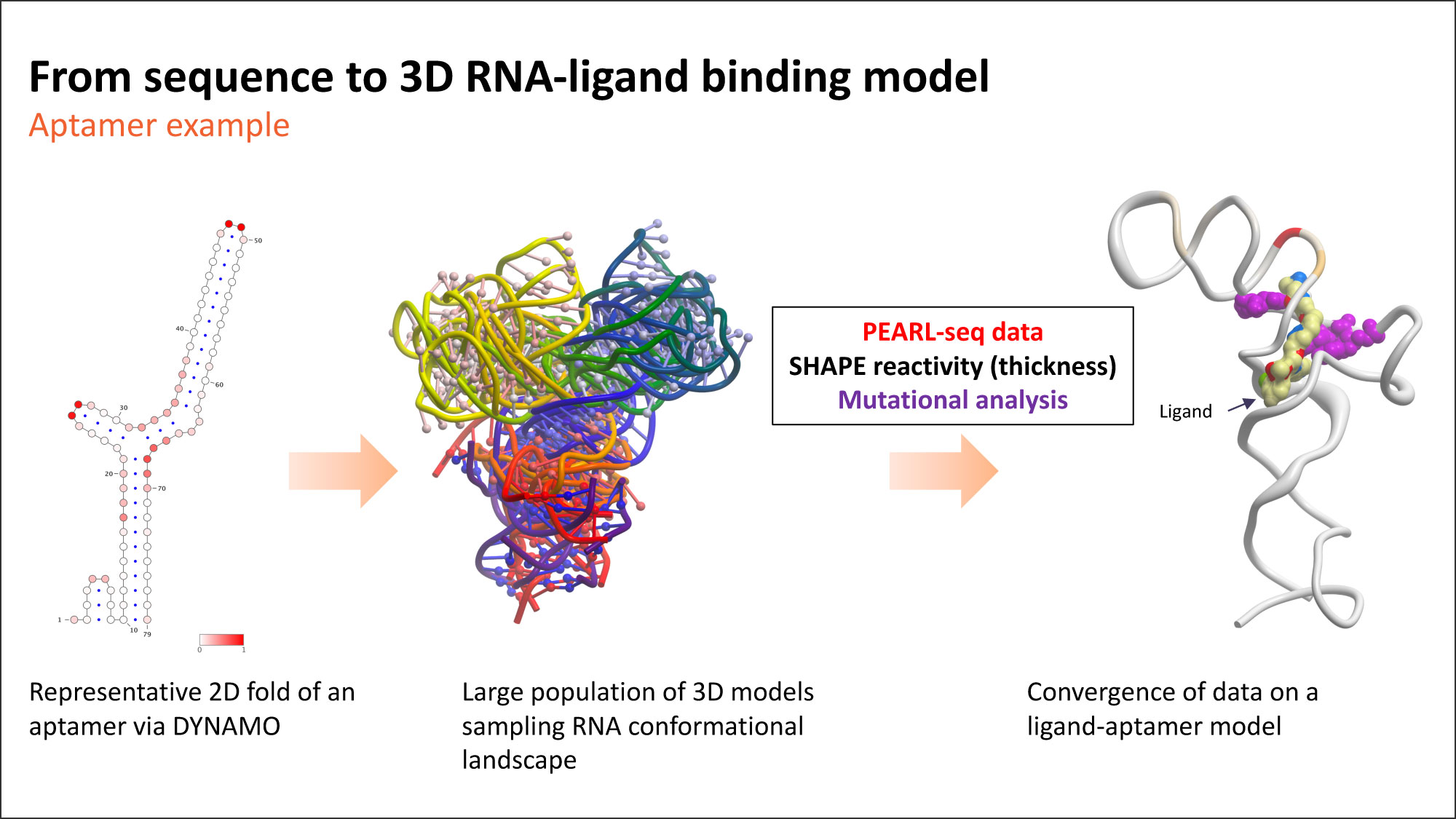
The central role of RNA’s cellular dynamics suggests this approach has great promise. Both experimental and computational evidence shows that RNA, in its biologically relevant forms, is often not in its lowest energy state. Rather, it’s in an excited state. That’s the state we must find to properly target these molecules. We are continually learning about the nuanced dynamics of RNA, but we are also excited by what we learn. Stay tuned…
