Reaching cruising altitude: New discovery tools to target RNA
Herschel Mukherjee, Scientist II, Chemistry, Arrakis
Craig Blain, Associate Director, RNA Biochemistry, Arrakis
 The majority of small molecule drugs induce their therapeutic effects by seeking out and binding to their intended target while avoiding most other molecules in the dense milieu of the cell interior. In doing so, biological processes that rely on a specific target can be selectively modulated, ultimately producing the desired therapeutic effect. Understanding the ability of a given drug candidate to bind its target, as well as identifying potential off-target interactions, is thus a critical part of the drug discovery process. However, since most small molecule drugs target proteins in the cell, the vast majority of target identification methods available to research scientists are aimed at identifying interactions between small molecules and proteins.
The majority of small molecule drugs induce their therapeutic effects by seeking out and binding to their intended target while avoiding most other molecules in the dense milieu of the cell interior. In doing so, biological processes that rely on a specific target can be selectively modulated, ultimately producing the desired therapeutic effect. Understanding the ability of a given drug candidate to bind its target, as well as identifying potential off-target interactions, is thus a critical part of the drug discovery process. However, since most small molecule drugs target proteins in the cell, the vast majority of target identification methods available to research scientists are aimed at identifying interactions between small molecules and proteins.
Our overall mission at Arrakis is to expand the set of “druggable” targets for small-molecule medicines to include RNA. But as our CEO, Mike Gilman, likes to remind us, “we’re building the plane as we’re flying it.” In other words, while our ultimate goal is to develop RNA-targeting small molecules (rSMs) which will provide a therapeutic benefit to patients, we also have to build the tools and platforms that will allow us to know that our molecules are doing what we want them to. Today, we are pleased to announce that our article describing one such platform: “PEARL-seq, A Photoaffinity Platform for the Analysis of Small Molecule-RNA Interactions” was published in ACS Chemical Biology.
Defining the flight path for PEARL-seq
Just as one wouldn’t step off a cliff with nothing but 200 tons of aluminum and steel, we weren’t starting from scratch either. As we peered over the edge, we identified three key questions that our hypothetical platform would address. First, given an RNA target, could we identify where on the RNA an rSM bound? Next, if we introduced an rSM into a mixture of RNAs, could we identify additional, “off-target” RNAs that were bound by our rSM? Finally, if we found an rSM that had biological activity, could we correlate that activity with the ability of the rSM to bind RNA? The initial discussions that would lead to the PEARL-seq platform involved scanning the literature for the wings, propellers, and engines that we thought we could use. Early work in the fields of chemical biology and RNA biology provided us clues about how we might achieve lift-off. But our aspirations required refining, combining, and expanding the building blocks that had previously been reported – we wanted a general-purpose technology, from probes to data analysis, to identify the RNA targets and binding sites of a compound across the full transcriptome.
PEARL-seq takes off
First, however, we needed a model system – a wind tunnel, if you will – to ensure that the various components of the PEARL-seq platform worked as intended. To this end, we selected a known rSM-RNA system, Aptamer 21, which possessed several attractive features. Among these was the presence of a flexible “linker” extending from the drug-like rSM, which would serve as the handle to which our photoactivatable warheads would be attached. After synthesizing a small panel of probes, we tested their ability to photocrosslink to the RNA using a gel-based assay. In this assay, the RNA was incubated with the various probes and then exposed to UV light. Whereas the probe itself binds reversibly with the RNA, UV light generates a highly reactive – but short-lived – intermediate that forms an irreversible bond with the RNA. Sites where the probes crosslinked to the RNA could then be determined by reverse transcription (RT) – RT enzymes typically cannot “read through” the bulky photoadduct and thus affords a truncated cDNA product. This assay yielded our first hint of success – two faint, but distinct, bands on an RT pausing gel using the probe that would later become “2c” in our publication.
Our expedition gains altitude
After that initial result, it was all hands on deck. Emblematic of Arrakis’ collaborative approach to science, almost all of the scientists in the lab—or, in the case of our colleagues in computational chemistry and biology, on their computers—at the time contributed to developing the PEARL-seq platform. As we continued to optimize our PEARL-seq probes and by testing them using the RT pausing gel assay, we also adapted the assay for next-generation sequencing analyses. Although the gel analysis was a convenient way to test our probes, it did not have the throughput or sensitivity required to scan the whole transcriptome for rSM-RNA interactions. We needed to turn our single propeller puddle-jumper into a jumbo jet.
We stuck with our trusty aptamer system, but instead of running the DNA products made by RT on a gel, we attached them to adapters that enabled next-generation sequencing. With millions of DNA sequences, we could precisely measure the rate of RT pausing at each position of the RNA. In agreement with our gel results, we found strong drop-off signals for the probes that depended on the rSM-RNA interaction.
PEARL-seq cruises to the transcriptome
It was now time to take PEARL-seq to the transcriptome! Searching for probe adducts through all the RNA in a cell is a daunting task. Fortunately, we’re not the first ones to tackle this challenge. RNA biologists have searched the transcriptome for protein interactions, residue modifications, structural motifs, and other features, giving us a validated set of sequencing tools to work with. Building on these methods, we were able to specifically recover the aptamer RNA from a pool of human mRNA, demonstrating our ability to pick out a specific rSM-RNA interaction from a complex mixture of RNAs and even to identify the same RT drop-off site that we saw in the targeted experiments. Analyzing these data required a sophisticated bioinformatics pipeline (the term for this specialized software), and our computational biologists worked alongside the wet lab scientists from the start to develop one. We built this pipeline in part on open source software created by the bioinformatics community, and, in turn, we have provided the source code for our pipeline along with the publication.
Guiding us on the journey ahead
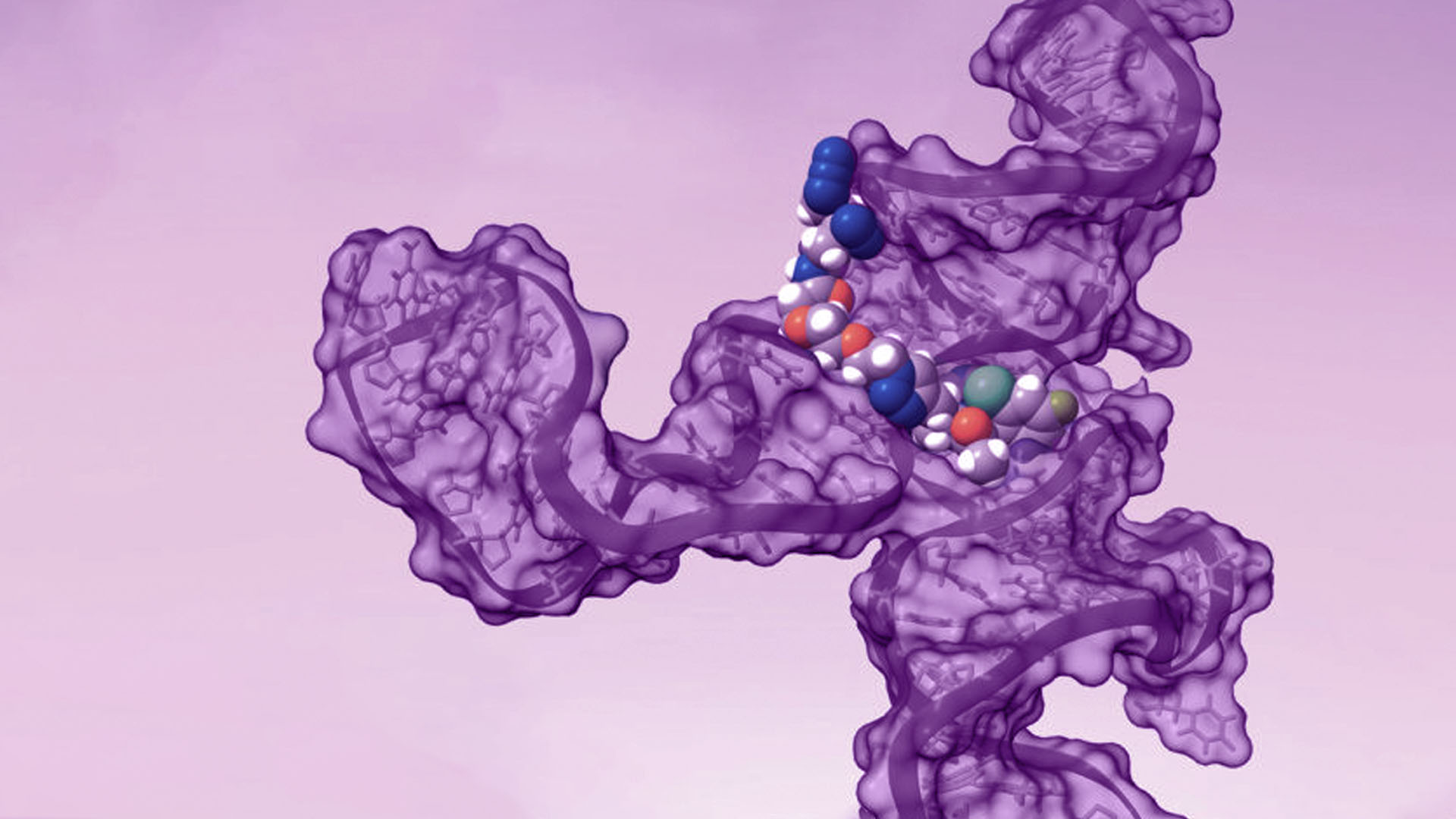
Finally, we showed how PEARL-seq can be combined with screening and structural analysis techniques to gain a fuller understanding an rSM-RNA interaction. Enlisting even more of our colleagues at Arrakis, we used SHAPE and DMS probing to refine our understanding of the secondary structure of our model aptamer. We used these 2D structures to predict an ensemble of possible 3D structures using MC-Sym software, and then filtered the structures for those containing potential binding pockets. Finally, we virtually docked our probe compound into these pockets to arrive at a model of the rSM-RNA structure that matched the PEARL-seq crosslinking data and the rSM structure-activity relationships. Again, highlighting the importance of collaboration to science at Arrakis we discussed some of our ideas with Prof. David Chenoweth, an Arrakis SAB member who leads a research group that is interested in developing new small molecule approaches for selectively modulating nucleic acid structure and function. While our publication focuses on a model Aptamer, similar models will be invaluable for us moving forward, as they will facilitate the design of rSM therapeutics in a rational manner, even in cases where high-resolution experimental data on RNA structure is unavailable.
Settling in for the long haul
The PEARL-seq platform is one of our instrument clusters – it can provide us with a lot of critical data on our journey towards rSM therapeutics. Not only can we demonstrate that our rSMs are engaging our target of interest, we can also scan for potential off-target RNAs and try to steer around them. In addition, by integrating PEARL-seq with our other platforms, we can generate model structures of our RNA targets, which will help us rationally design new therapeutics. We’ve still got a long way to go at Arrakis, but the PEARL-seq platform represents a significant milestone for our science. With any luck, we’ll figure out our landing gear next.
